Take a deep breath: fast and accurate lung segmentation using conventional image processing
Automation of lung segmentation and medical image analysis has been explored for many years. With the rapid development of medical imaging, the volume of medical data being stored has seen exponential growth. Nowadays, there are over 600 million advanced medical imaging procedures per year – and this is just in the US. This sums up to 1 EB (1 million terabytes) of total U.S. medical storage and archiving data as of 2016 [1]. An average CT scan of lungs consists of hundreds of horizontal cross-sections (we call them slices) and takes several hundred megabytes of storage space.
What is lung segmentation?
Lung segmentation is a process of partitioning an image into lung and non-lung segments. To put it simply, given an image, we want to select only the regions or segments occupied by lungs. Additionally, the lungs can be further segmented into anatomical regions such as bronchi, lobes etc. In the case of an X-Ray, the segments consist of pixels. In Computed Tomography (CT), the regions consist of voxels, as they are three-dimensional. The process can be either manual, semi-automatic or fully automatic.
Before development of algorithms which automated the process, the procedure was performed by radiologists and it could be very time-consuming. With state of the art algorithms, the segmentation can take as little as few seconds. While manual segmentation can take dozens of minutes. Moreover, even several hours depending on the required precision. Lung segmentation is used mainly as a preliminary step for further analysis. Its applications range from very precise measurement of lung capacity to surgery planning and further segmentation of lesions – i.e., lung cancer.
Speed and precision in medical image segmentation
The reason why, to a large extent, the segmentation is currently performed by machines and not by humans is simple – machines are much faster. The obvious concern is whether the algorithms are also as precise as humans and the answer in most of the cases is yes. Even when the algorithm produces slightly erratic results, their correction is much faster than segmenting the whole scan manually. A fully automatic segmentation without any kind of human supervision is currently impossible because the algorithm cannot make sense out of the data it is processing contrary to a trained physician.
In most of the cases however, the results are quite precise, often more detailed than human-produced segmentation. The additional bonus is that algorithms are not susceptible to both inter- and intra-operator error. In other words, when a human is given the same study to segment several times, each segmentation will differ slightly. It is even more evident when several physicians are given the same study to work on. On the other hand, an algorithm will always produce the same results for the same study, regardless of how many times the procedure is repeated.
Why does it have to be so fast?
When one realizes the already mentioned volume of medical imaging data being stored, it becomes obvious that manual segmentation is no longer an option. Especially when additional constraints are put on costs and the number of trained personnel. Provided that the high accuracy is maintained, the faster the data can be processed the better. This also means that the patient can get the diagnosis much faster than ever before and humans no longer have to perform very tedious and repetitive tasks – instead, they can focus more on verifying the results and diagnosis. Furthermore, much of the already stored data is analyzed only in terms of the original goal of the examination. In the future, its automatic analysis, thanks to its speed and precision, could lead to new discoveries and reveal phenomena we are currently unaware of.
Lung segmentation – how is it achieved?
Algorithms for lung segmentation can be based on machine learning, traditional image processing methods, or even a mix of both. Machine learning is often used when lesions have to be segmented or when lung regions contain pathologic changes such as cancer [2]. The main disadvantage of machine learning is that the results strongly depend on the quality of the training set and in the case of medical imaging, it is still hard to obtain large, pre-labelled training sets – mainly due to ethical/legal reasons.
For healthy or slightly abnormal regions, very high accuracy can be achieved using conventional image processing such as image morphology, thresholding etc. for segmenting lung surface and bronchial tubes [3][4]. ECONIB team recently developed an algorithm that segments lungs with high speed and accuracy [4] reaching 11ms/slice processing time and 0.97 F1-Score with respect to human-segmented ground truth. The block diagram of the algorithm is rendered in Fig. 1.
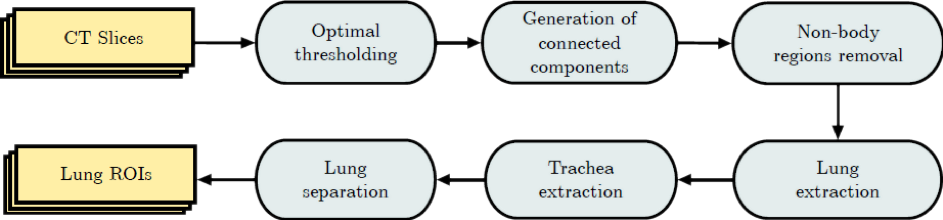
Fig. 1. – Block diagram of the algorithm developed by the ECONIB team
Algorithm development – the process
First, the image is thresholded – the white pixels correspond to lung tissue and air-filled regions and the black ones cover the remaining regions of the image (Fig. 2a). Next, in order to discard the thresholded regions which are not lungs, a set of 3d connected components is constructed to check how they connect in space. The large connected region at the bottom is the bed occupied by the patient and it has to be removed (Fig. 2b, bed marked green). It can be safely assumed that the largest remaining air-filled region will be within the patient’s body and it spans the whole lungs. This however is not the end – the region also contains other air-filled areas – trachea and upper airways (Fig. 2c, upper airways marked green) which have to be removed.
This is achieved using the trachea extraction. This is the step which moves a planar cross-section from the top of the patient’s head to the bottom. Then the area grows too rapidly, the region is detached. Such growth occurs for the first time when trachea connects with the lungs. This way, the upper airways, together with the trachea, can be removed. In the last step, using a combination of morphological operations, the lungs which often appear to be connected at sternum due to insufficient image resolution can be separated. This is achieved using a combination of repetitive morphological operations such as erosion and dilatation. Finally, the remaining black and white masks contain only the lungs. Furthermore, they are separated so that they can be automatically labeled as left and right lung (Fig. 2d).
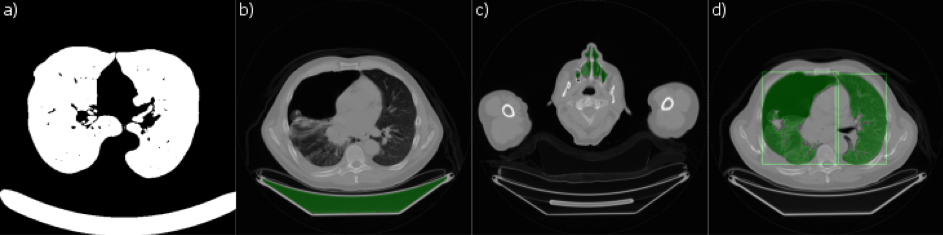
Fig. 2. – Some of the intermediate steps of the lung segmentation algorithm
Algorithm development – the result
As the result, a very fast and efficient algorithm relying solely on domain knowledge and conventional computer vision techniques was built. The main advantage compared to machine-learning based approaches is that it does not require training at all and it achieves high speed and accuracy. The final result rendered in 3D is presented in Fig. 3. Note the high level of details in the segmented image. Detailed structures such as bronchial tubes inside the lungs are visible. The segmented surface is visibly deformed by the ribs.
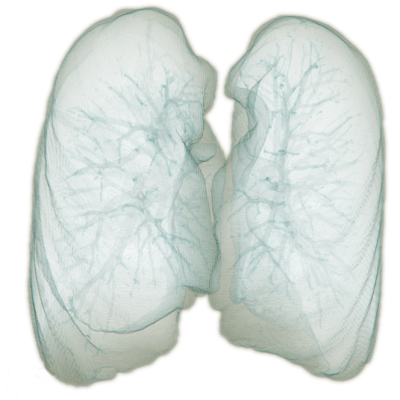
Fig. 3. – 3D rendering of the lung segmentation produced with the algorithm developed by ECONIB
Bibliography
[1] Storage Capacity Management For EMR – Electronic Medical Records (PACS), https://turbonomic.com/blog/on-technology/storage-capacity-management-emr-electronic-medical-records-pacs/
[2] Schlegl, T., et al.: Unsupervised pre-training across image domains improves lung tissue classification. Medical Computer Vision: Algorithms for Big Data pp. 82-93 (2014)
[3] Costa, A., Carvalho, B.: SALSA – A simple automatic lung segmentation algorithm. Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications, pp. 501-508 (2015)
[4] Walczak M., Burda I., Nalepa J., Kawulok M., Segmenting Lungs from Whole-Body CT Scans, Beyond Databases, Architectures, and Structures, Communications in Computer and Information Science, vol. 427, pp. 403-414
Contact us if you have any questions!
Read more about our AI measurements