Shedding Light on the Black Box: A Gradient-Based Approach to Explainable AI in the Medical Field
The Explainable Artificial Intelligence
Artificial Intelligence (AI) and Machine Learning (ML) models are often considered black boxes. We train them, validate their performance on test sets, and analyze their effectiveness. However, sometimes we need a deeper understanding of model predictions. In a medical field, the key question arises: which areas of the input image had the greatest impact on the model’s final decision?
To answer this, we use Explainable Artificial Intelligence (XAI) methods, which provide insights into the model’s decision-making process.
When to use XAI
Certain machine learning tasks, such as classification, are well-suited for XAI methods. For example, given an image of a skin lesion, the model classifies it as cancerous or non-cancerous. In such cases, understanding which regions influenced the classification decision can aid in model validation.
Image classification is relatively straightforward since finding relevant regions is easier. However, tasks like segmentation require each pixel or voxel to be classified independently. XAI helps identify regions that should have been segmented but were ignored due to the network’s limitations, such as insufficient feature or activation mapping.
XAI methods
There are several XAI techniques. One of the most widely used is the model-agnostic method LIME (Local Interpretable Model-Agnostic Explanations) [1]. LIME perturbs input data and observes its effect on the target network, forming an explainer based on a simpler model such as a decision tree.
Despite its advantages, LIME must be carefully handled. The nature of input perturbations should be known in advance to ensure proper network learning.
In this article, we focus on gradient-based methods such as Grad-CAM [2] and Grad-CAM++ [3], which are more effective for XAI across various tasks.
Gradient-based XAI methods
Grad-CAM and Grad-CAM++ are powerful gradient-based visualization techniques. Both methods rely on feature maps from the last convolutional layer. The saliency map is computed as a weighted sum of these feature maps, where the weights are based on the partial derivatives of the target class with respect to the feature maps.
\[ L^c_{ij}= \sum_k w^c_k A^k_{ij} \]
where
– \(L^c_{ij}\) is the class-specific localization map,
– \(A^k_{ij}\) is the feature map from the last convolutional layer (for pixel i,j),
– \(w_k^c\) are the weights computed as:
\[w^c_k= \frac{1}{Z}\sum_i \sum_{j} \text{ReLu}\left( \frac{\partial y^c}{\partial A^k_{ij}}\right)\]
where:
– \(Z\) is a scalar normalization factor,
– \(y^c\) is the model’s output score (logit) for class c, before applying activation function
Grad-CAM++ extends this by incorporating higher-order derivatives, refining the focus on multiple relevant objects. The weight computation in Grad-CAM++ is modified as:
\[w^c_k= \sum_i \sum_j \alpha_{ij}^{kc}\text{ReLu}\left( \frac{\partial y^{c}}{\partial A^k_{ij}}\right)\]
Where:
\[\alpha_{ij}^{kc} = \frac{\frac{\partial^2 y^c}{\partial (A^k_{ij})^2}}{2 \frac{\partial^2 y^c}{\partial (A^k_{ij})^2} + \sum_{m,n} A^k_{mn} \frac{\partial^3 y^c}{\partial (A^k_{mn})^3}}\]
where higher-order derivatives help distribute importance more effectively across multiple objects.
Comparison of grad-cam methods
Both methods generate visual representations of activation maps, highlighting important regions in the input image. Grad-CAM++ improves upon Grad-CAM by offering more precise localization. For example, when analyzing images with multiple objects of the same class, Grad-CAM may focus on only one object, while Grad-CAM++ distributes attention across all relevant objects.
Below, we present results from medical segmentation tasks using both methods [4] :
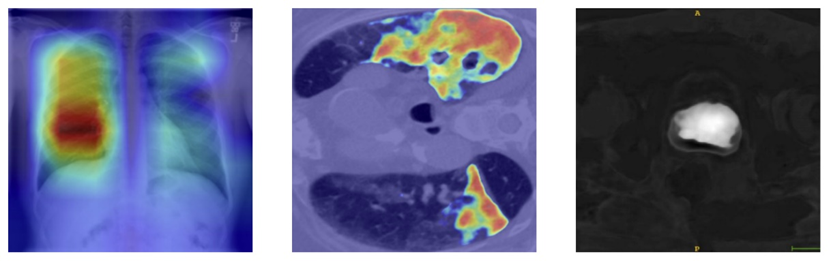
Figure 1. Grad-cam results for a) Chest X-ray for classification, b) Lung CT slice for 2D segmentation, c) Prostate CT scan for 3D segmentation
Grad-cam++
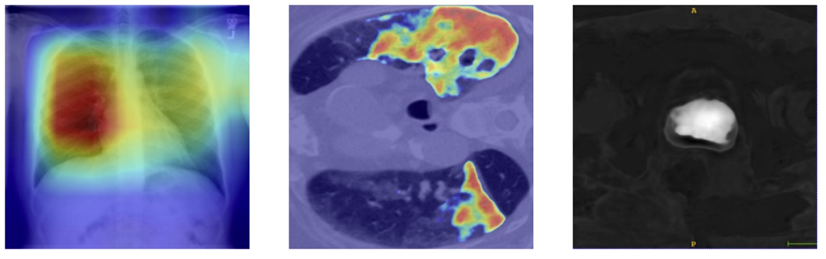
Figure 2. Grad-cam++ results for a) Chest X-ray for classification, b) Lung CT slice for 2D segmentation, c) Prostate CT scan for 3D segmentation
Conclusions
Gradient-based XAI methods are gaining traction, with numerous implementations available. Some networks even come with built-in XAI modules. For further reading, we recommend exploring M3D-CAM, a tool that integrates seamlessly with most convolutional networks.
References:
- M. T. Ribeiro, S. Singh, and C. Guestrin, “Why should i trust you?: Explaining the predictions of any classifier,” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM, 2016, pp. 1135–1144.
- R. R. Selvaraju, A. Das, R. Vedantam, M. Cogswell, D. Parikh, and D. Batra, “Grad-cam: Why did you say that? visual explanations from deep networks via gradient-based localization,” arXiv preprint arXiv:1610.02391, 2016
- Aditya Chattopadhyay, Anirban Sarkar, Prantik Howlader,and Vineeth N Balasubramanian ,Grad-CAM++: Improved Visual Explanations for Deep Convolutional Networks, 2017, https://doi.org/10.48550/arXiv.1710.11063
- Karol Gotkowski, Camila Gonzalez, Andreas Bucher, Anirban Mukhopadhyay, M3d-CAM: A PyTorch library to generate 3D data attention maps for medical deep learning, 2020, https://doi.org/10.48550/arXiv.2007.0045