Mamba Rising: Are State Space Models like U-Mamba Going to Replace Ordinary U-Net?
The U-Net has been the gold standard for medical image semantic segmentation for years. While its elegant architecture remains effective, its success has led to an observable plateau. In fact, the nnU-Net framework [1], which represents the current state-of-the-art approach for training the U-Net, consistently achieves extremely high performance, setting a difficult benchmark for competitors.
The Problems with U-Net Successors
Many new models are merely incremental variations of a classic U-Net, like UNet++ [2] or Attention U-Net [3], offering only marginal gains in specific tasks. Furthermore, new architectures often do not make the grade when tested in emerging works. Simple enhancements to the U-Net’s training regime frequently allow it to achieve the same improvements claimed by these supposedly superior models. On the other hand, transformer-based approaches like UNETR++ [4] are notoriously resource-intensive, requiring large data volumes and expensive GPUs with substantial VRAM. Critically, they also share the same fundamental issue as CNNs: they often fail to demonstrate lasting superiority over improved U-Net baselines in subsequent testing. This highlights a clear need: the field requires a novel architecture that offers a significant leap in performance and is not dependent on large data volumes. This is precisely the gap that U-Mamba [5] aims to fill.
What are State Space Models (SSMs)?
The search for efficiency in deep learning has led to the exploration of new architectural paradigms like State Space Models (SSMs) [6]. The motivation for SSMs comes from a key weakness in transformers: their self-attention mechanism scales quadratically. This means that as an image or sequence gets larger, the computation required explodes. SSMs are an entirely different approach. The promise of SSMs is linear scaling. They are designed to process long sequences of data with incredible efficiency. To put this in perspective: Imagine you want to double your patch size to capture more context. For a transformer, this 2x increase in input size causes the memory and compute cost to surge by 4x (22). In contrast, for U-Mamba, the cost simply doubles.
SSMs originate from control theory and the mathematical modelling of physical systems, often using linear time-invariant (LTI) concepts. Unlike transformers, which calculate global interactions through parallel attention weights, SSMs process information sequentially. At their core, they maintain a compressed “state” of all the information seen so far, updating it with each new piece of data. The key innovation in Mamba is a selection mechanism. This “gate” intelligently decides what information to keep in its state and what to forget. This reliance on a hidden state, rather than a global attention matrix, is the fundamental difference from the transformer architecture. Crucially, unlike recurrent neural networks, SSMs are structured such that the state computation can be transformed into a convolution, enabling massive parallel processing during training.
In a medical context, this is powerful. We can treat an image as a long sequence of pixels. An SSM can “see” a pixel in the top-left corner and, thousands of pixels later, remember its context when analysing the bottom-right corner. This allows it to model long-range dependencies. For a radiologist, this is like understanding the subtle relationship between a small lesion in one lobe of the lung and a faint pattern in another. This global understanding is something traditional U-Nets can struggle with.
U-Mamba architecture
The U-Mamba architecture is a hybrid model, and its motivation is very clever: don’t reinvent the wheel, just improve it. The U-Net is brilliant at learning local features – edges, textures, and small shapes. Its skip-connection structure is unmatched for preserving fine-grained spatial detail. U-Mamba does not throw this away. Instead, it augments the U-Net by replacing some standard convolutional blocks with the new U-Mamba Block.
The promise here is to get the best of both worlds. As seen in the architecture diagram, the model uses the familiar U-Net encoder-decoder skeleton. The convolutional layers capture the “what”, e.g., “this texture looks like a cell”. The U-Mamba blocks, integrated within, provide the “where” and “why”, e.g., this cell’s relationship to the entire tissue structure suggests it’s anomalous. It is a synergistic design where convolutions handle the local details, and Mamba handles the global context. This allows the model to “think” more like an expert, using broad context to inform local decisions.
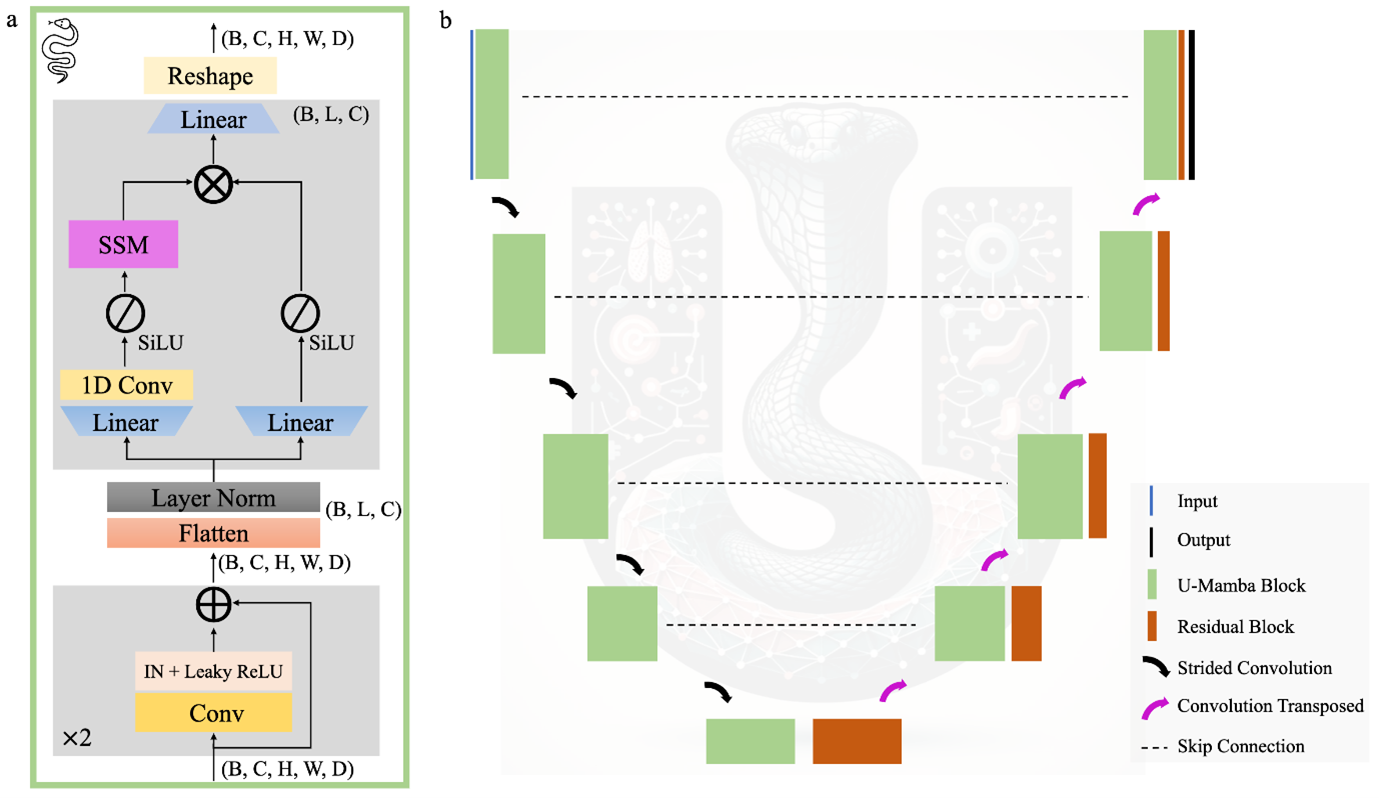
Figure 1. Overview of the key features of the U-Mamba architecture. (a) U-Mamba Building Block: This block is composed of two successive residual blocks followed by the Mamba block, which is included to enhance long-range dependencies. (b) U-Mamba Encoder Architecture: This illustrates the overall architecture of the U-Mamba Encoder configuration, where the U-Mamba block is included in all blocks building the encoder. Figure sourced from [5].
To achieve this, the U-Mamba focuses on three key functional components:
- Sequential modelling: The core State Space Model within the U-Mamba Block is responsible for processing data as a sequence, efficiently modelling long-range dependencies within the feature map.
- Dynamic gating: The block utilizes a complex gating mechanism (including a linear layer, Sigmoid Linear Unit (SiLU) activation, and multiplication) to dynamically modulate the flow of information based on the input.
- Spatial integrity: The overall U-Net structure retains its defining features: strided convolutions in the encoder for efficient downsampling and transposed convolution layers in the decoder for precise upsampling. Crucially, the skip connections are retained to ensure that fine, high-resolution details are passed directly across the model.
U-Mamba can be run in two configurations. The first, where the U-Mamba block is used solely in the bottleneck, is called U-Mamba Bottleneck. The second, where the U-Mamba block is used in all encoder blocks, is called U-Mamba Encoder. While U-Mamba Bottleneck performs slightly worse on some tasks, the benefit of including the U-Mamba block in the bottleneck only is that it is significantly less memory intensive.
U-Mamba: A Plug-and-Play Solution
Beyond its novel architecture, one of U-Mamba’s greatest features is its practicality. It is not just a theoretical paper; it is designed to be plug-n-play. The installation is straightforward and involves only a few simple `pip install` commands. The decision to build it directly on the nnU-Net framework was a critical one. This ensures a fair comparison against the reigning standard. For researchers, this is invaluable. It means you are testing the architecture itself, not just a specific, lucky implementation of training regime. Because U-Mamba uses the same data format as nnU-Net, teams can skip the time-consuming preprocessing step if it was already performed. This low-friction setup allows teams to leverage their existing nnU-Net pipelines, using the same data formats and preprocessing steps, which drastically lowers the barrier to adoption. You may, in the worst case, only need to modify the `plans.json` file.
Hands-On Experience
Our internal experiments provide a practical, hands-on verdict. The answer to the question in the title is clear: U-Mamba is not yet going to replace U-Net, but it is the most viable challenger we have seen to date. We found that U-Mamba is not a solution that always prove to be superior U-Net on all tasks. However, it demonstrated significant advantages in specific, challenging areas. For instance, it performed exceptionally well when segmenting very small objects, likely because it could use global context to find them. In abdominal organ segmentation (see Fig. 2), U-Mamba demonstrated more accurate segmentation masks for the liver and stomach in CT scans, and the gallbladder in MRI scans, relative to established U-Net and Transformer baselines. Furthermore, U-Mamba training dynamics were impressive. U-Mamba often achieved good, stable segmentation earlier in the training process and did not require as much heavy-hand tuning (like custom loss functions or massive patch sizes) that nnU-Net sometimes needs to reach its peak. The promises are largely fulfilled. It has a clear trade-off: it requires more VRAM than U-Net (U-Mamba Bottleneck about 2x), but it is still far more accessible than a full transformer model. It represents a powerful, new tool in our developer toolbox.
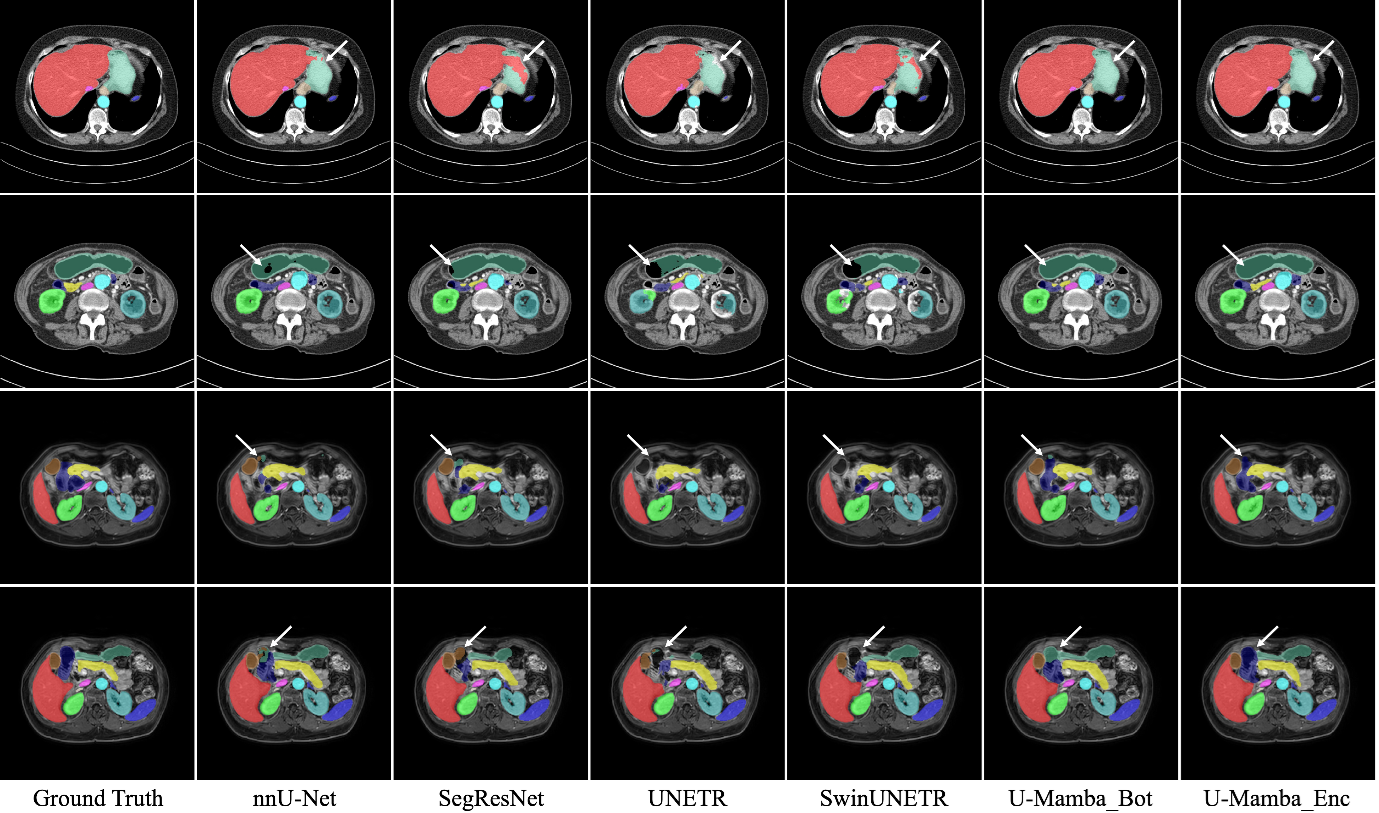
Figure 2. Comparison of segmentations examples for U-Mamba and predecessor models. This figure provides visual results for abdominal organ segmentation on CT (1st and 2nd rows) and MRI scans (3rd and 4th rows). Figure sourced from [5].
Resources:
[1] Isensee, F. et al. (2024). nnU-Net Revisited: A Call for Rigorous Validation in 3D Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2024 (pp. 488–498). Springer Nature Switzerland.
[2] Zhou, Z., Siddiquee, M., Tajbakhsh, N., & Liang, J. (2019). UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Transactions on Medical Imaging.
[3] Oktay, O. et al. (2018). Attention U-Net: Learning Where to Look for the Pancreas. arXiv preprint arXiv:1804.03999.
[4] Shaker, A., Maaz, M., Rasheed, H., Khan, S., Yang, M.H., & Shahbaz Khan, F. (2024). UNETR++: Delving Into Efficient and Accurate 3D Medical Image Segmentation. IEEE Transactions on Medical Imaging, 43(9), 3377-3390.
[5] Ma, J., Li, F., & Wang, B. (2024). U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation. arXiv preprint arXiv:2401.04722.
[6] Gu, A., Johnson, I., Goel, K., Saab, K., Dao, T., Rudra, A., & Re, C. (2021). Combining Recurrent, Convolutional, and Continuous-time Models with Linear State-Space Layers.